The use of integrative taxonomy methods in the delimitation of lichen species on the example of the critical genus Wetmoreana (Teloschistaceae, lichenized fungi)
Summary
DNA sequencing methods were introduced into lichenology quite recently before the turn of the millennium. They resulted in a revolution in the classification of lichens and the delimitation of species. Molecular analyses revealed that the species richness of lichens is grossly underestimated. A part of this unrecognized diversity are so-called cryptic species, which are distinct in terms of their genetic makeup but morphologically similar. Many lichen groups, including the family Teloschistaceae, also include cryptic genera such as Caloplaca in a broad sense, which consists of morphologically similar but unrelated species. Modern taxonomy takes an integrative approach, by incorporation DNA sequencing, morphology, biochemistry, ecology, and biogeography. However, in the case of the family Teloschistaceae, recent adjustments in the classification of genera largely rely on DNA sequence data only.
The aim of the present project was to learn novel statistical methods used in integrative taxonomy for the delimitation of genera and species in Teloschistaceae, focusing on taxa previously included in a broadly defined Caloplaca. As a case study, we selected the genus Wetmoreana, mostly known from South America. The research was conducted as part of a scientific internship under the supervision of Dr. Robert Lücking, at the Botanic Garden and Botanical Museum of the Freie Universität, Berlin, Germany.
The employed novel method is called phenotype-based phylogenetic binning. It uses a reference tree of phylogenetic relationships based on DNA sequences for a set of reference taxa. For the same taxa, a matrix of phenotype characters is established and the phylogenetic signal of each character is then evaluated by analyzing their distribution on the tree through a weighting approach. Additional specimens for which no DNA sequences are available can then be binned into the reference tree by computing their most likely placement based on the weighted phenotype characters. Possible alternative placements are thereby statistically evaluated through so-called bootstrapping, a random analysis of a subset of characters. In addition, a method called the multiple response permutation procedure was used to determine whether groupings based on DNA sequence data are also distinguished by phenotype characters. For these approaches, programs such as RAxML and PC-ORD are available.
In the present case, we evaluated the possible placement of certain species within the genus Wetmoreana, based on a matrix of 65 characters for 116 specimens. This newly learned methods of integrative taxonomy will be used in an expanded study on representatives of the family Teloschistaceae in South America. This group of lichens is very poorly investigated in that continent, and both traditional taxonomic treatments and phylogenetic studies are scarce. At the same time, the systematics of Teloschistaceae, which is one of the most species-rich groups of lichens (contains about 1,000 species), is currently based almost exclusively on phylogenetic DNA sequence data. As a consequence, it is impossible to classify species for which sequence data are not available. The phylogenetic binning method may help to solve this problem, as well as shed light on current taxonomic approaches based on molecular data. The preliminary results of the study conducted on representatives of the genus Wetmoreana (Teloschistaceae family) from South America are very promising. This study is an innovative approach, previously not practiced for the family under investigation.

Wetmoreana sp. nov. – squamulose thallus with abundant apothecia and pycnidia.
Photo: Karina Wilk.
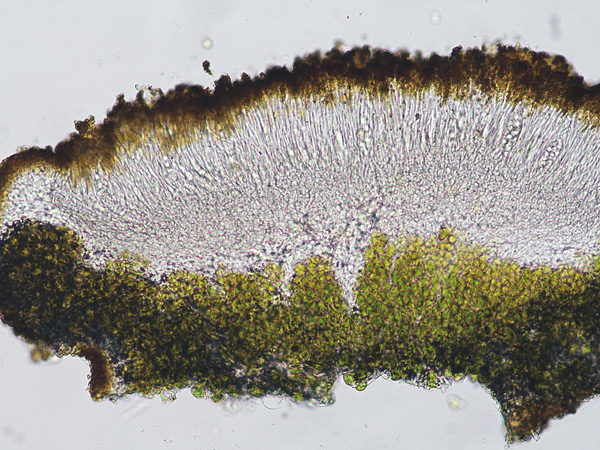
Wetmoreana sp. nov. – apothecia zeorine with proper and thalline margins.
Photo: Karina Wilk.
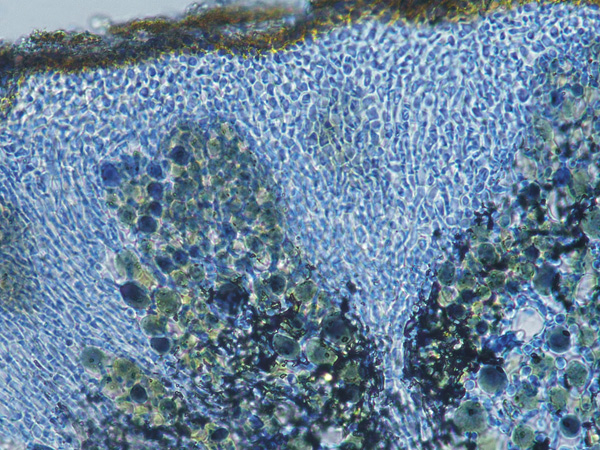
Wetmoreana sp. nov. – structure of the thalline cortex in Lacto-phenol Cotton Blue.
Photo: Karina Wilk.